Technical Support技术支持
CONTACT US
 400 179 0116
400 179 0116
24-hour service hotline marketing@ldraft.comE-mail
marketing@ldraft.comE-mail
Key Points - Essential for Cell Knockout
source:QiDa technoligy views:2203 time:2023-04-27
Taking a path that is rarely traveled, producing a knockout cell line instead of knockout cells? In this article, we introduce you to techniques and tricks for utilizing homologous directed repair pathways, designing optimal donor DNA, and avoiding common accidents in genome editing.
Knock in mutation and genome editing
Targeted genome editing events can be divided into two categories: cell knockout and animal knockout. The purpose of knockout is to disrupt DNA translation by generating frameshift mutations. Most cell repair events generate these types of mutations, making it a "simple" editing task. However, for knocking bricks, this is not the case. Knocking in mutations usually requires binding to precise DNA sequences at precise genomic locations, with almost no margin for error. Not surprisingly, these editors require more planning and skills than eliminating opponents.
To start a knockout experiment, the first question to ask is where to knock out, so you need to determine the location of the knockout brick you want to introduce into the genome. Then choose which Cas enzyme to use and design a gRNA where you want to introduce editing. Finally, you must know what sequence to introduce, which is accomplished by designing and using donor DNA molecules. Your donor molecule needs to have homology with the target gene loci on both sides of the new sequence. This homology will guide cells to use donor DNA as a template for repair, which will allow your sequence to bind and ultimately knock out successfully! So, how are donors used as templates for repair? Please see the breakdown below.
Homologous directed repair and genome engineering
The simplest way to create knockout cell lines is to utilize the built-in repair pathways that cells already possess to repair DNA double strand breaks - (HR).
HR is the dominant homologous directed repair (HDR) pathway in mammalian cells, but it is usually a less common DNA repair mechanism. On average, it only accounts for less than 10% of DNA repair, and the repair rate varies among different cell types. The difference may be caused by cell cycle rate; Rapidly growing cells will have a higher HR frequency than slowly growing cells. It is important to know that HR often undergoes mutations in cancer cells, especially in commonly used breast and ovarian cell lines in the laboratory. We suggest that you check for HR related mutations in the cell line before attempting HDR knockout.
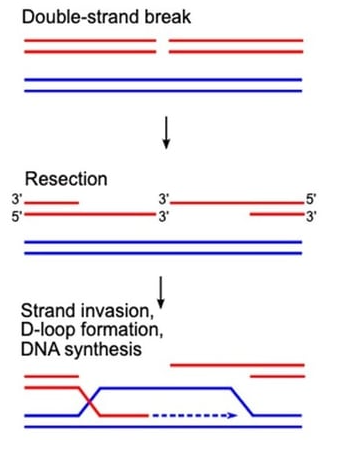
Figure 1: Early steps in repairing DNA double strand breaks through homologous recombination.
Design HDR
The basis of HDR pathway depends on the repair of template molecules, which are usually endogenous available sisters chromatids, although exogenous DNA can also be used. Here are some considerations for designing donor DNA for successful HDR activities.
CRISPR cutting position
Place the CRISPR cutting area as close as possible to the tapping position. HDR relies on cutting off the broken site to form ssDNA and using the end of ssDNA to find repair templates. When the insert is within 10bps of cleavage, the highest HDR efficiency is observed, making the cleavage essentially part of the template DNA.
Homologous sequences and sample types
To ensure that your donor molecule is used as a template, you need to add homologous sequences to the right and/or left of the knock in site. The length and composition of the sequence (single stranded and double stranded) depend on the size of the carrier. Single stranded oligonucleotide donors (ssODNs) with homologous sequences up to 40bps can effectively knock in sequences of several hundred bps. However, if you have a larger knockout (200 bp – 2 kb), due to the synthesis limitations of oligonucleotides, you will need to use dsDNA donors. These vectors traditionally have large homologous sequences in the range of 500bp to 1kb, but recently, shorter homologous regions have also been used with some success (Yu et al., Nat Chem Biol).
Mutated PAM or sgRNA sequence
Cas9 may undergo multiple rounds of cleavage at the target site until a portion of the PAM site or guide recognition sequence is disrupted through mutation. By introducing mutations in the PAM or guided recognition sequence of donor DNA, it can be ensured that HDR will not target again. If your Cas9 enters the coding region of a gene, please ensure that the PAM editing you introduce is a silent mutation, so that you will not accidentally change the protein you are interested in.
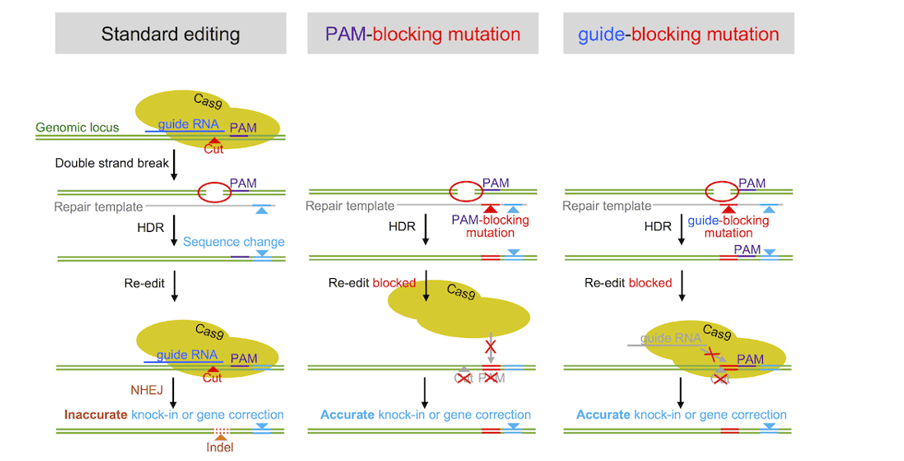
Figure 2: Stopping PAM re cleavage and guiding RNA disruption mutations in donor DNA
Elevating HDR over other editors
HDR is not the most common editing pathway for DNA repair. To tilt the scale during HDR editing, you can try one or more different editing positions.
Other repair mechanisms
In addition to HR, mammalian cells also have two main repair mechanisms - non homologous terminal junction (NHEJ) and alternative terminal junction (alt Ej). Inhibitors of important components of these two pathways have been shown to be effective in increasing HR editing success (Yang et al., Int J Mol Sci). Adding these inhibitors to the culture medium before and after the introduction of Cas9 will increase HDR, ensuring that the inhibitors are removed within one or two days to avoid affecting cell viability.
Enriched S-phase cells
HR mainly acts on the S phase and G2 phase phase of the cell cycle. At this time, sisters chromatids may exist as repair templates. To maximize HDR release, ensure that cells actively circulate during the cell cycle stage. If cells are over fused or there are not enough nutrients in the culture medium, they will not carry out DNA replication, so pay attention to these cells! Furthermore, you can suppress the cell cycle regulatory proteins responsible for S phase transition, allowing cells to stay longer in the HDR promoted cycle. If the cell residence time is too long, this method may have some negative effects on survival ability. Long term cell cycle arrest can lead to cell apoptosis in just 10 hours.
Using fast Cas editing
What if promoting human resources is as simple as replacing your Cas9? sure! Cas12a is a member of the Cas family and produces sticky ends during cutting. Cas12a may promote HDR by producing ssDNA suspensions at the fracture site, which is an early HR step (Moreno Mateos et al., Nat Com).
Another method to start Cas cleavage of HR is to fuse HR protein to nuclease. The early HR factors fused with Cas9 have been shown to increase HDR frequency by interrupting HR before other factors are loaded onto the fracture, which may lead to repair following different pathways (Charpentier et al., Nat Com).
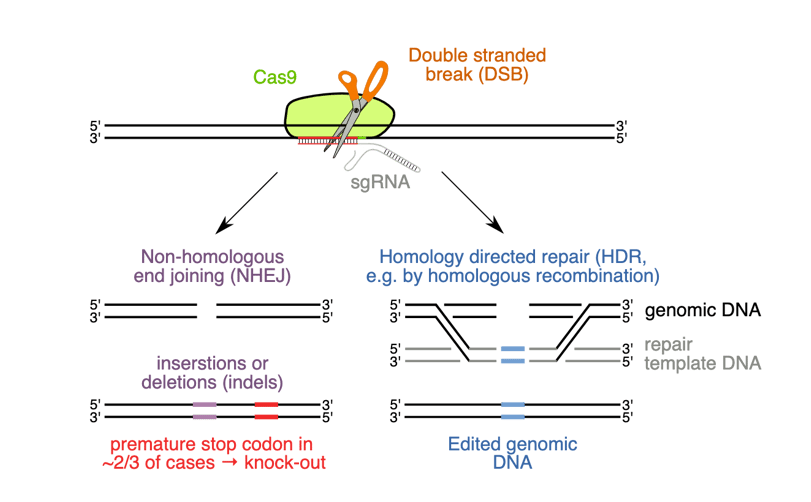
Figure 3: HDR entry competes with NHEJ mediated small insertion and deletion editing.
References
Yu, Y., Guo, Y., Tian, Qigi, Lan, Y., et. al. An efficient gene knock-in strategy using 5’-modified dsDNA donors with short homology arms. Nat Chem Biol., 308(20): 1-9 (2020). 10.1038/s41589-019-0432-1
Mehdi Banan. Recent advances in CRISPR/Cas9-mediated knock-ins in mammalian cells. J Biotechnol., 16(4): 387-390 (2020). 10.1016/j.jbiotec.2019.11.010
Wright, D. W., Shah, S. S., Heyer, W. D. Homologous recombination and the repair of DNA double strand breaks. J Biol Chem., 293(27): 10524-10535 (2018). 1 10.1074/jbc.TM118.000372
Yang, H., Ren, S., Yu, S., Pan, H., et al. Methods favoring homology-directed repair choice in response to CRISPR/Cas9 Induced-double strand breaks. Int J Mol Sci., 21(18): 6461 (2020). 10.3390/ijms21186461
Lin, S., Staahl, B. T., Alla, R. K., Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLIFE. (2014) 10.7554/eLife.04766
Moreno-Mateos, M. A., Fernandez, J. P., Rouet, R., Vejnar, C. E., et al. CRISPR-Cpf1 mediates efficient homology-directed repair and termperature-controlled genome editing. Nat. Com., 8(2024), (2017). 10.1038/s41467-017-01836-2
Charpentier, M., Khedher, A. H. Y., Menerot, S., Brion, A., et al. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat. Com., 9-1133 (2018). 10.3390/ijms21186461







